Contributors
- Sebastian Lindner (GitHub @Bienenwolf655; Twitter @lindner_seb)
- Núria Mimbrero Pelegrí (GitHub @nuriamimbreropelegri;)
- Michael Heinzinger (GitHub @mheinzinger; Twitter @HeinzingerM)
- Noelia Ferruz (GitHub @noeliaferruz; Twitter @ferruz_noelia; Webpage: www.aiproteindesign.com )
- Alex Vicente
REXzyme: A Translation Machine for the Generation of New-to-Nature Enzymes
Work in Progress
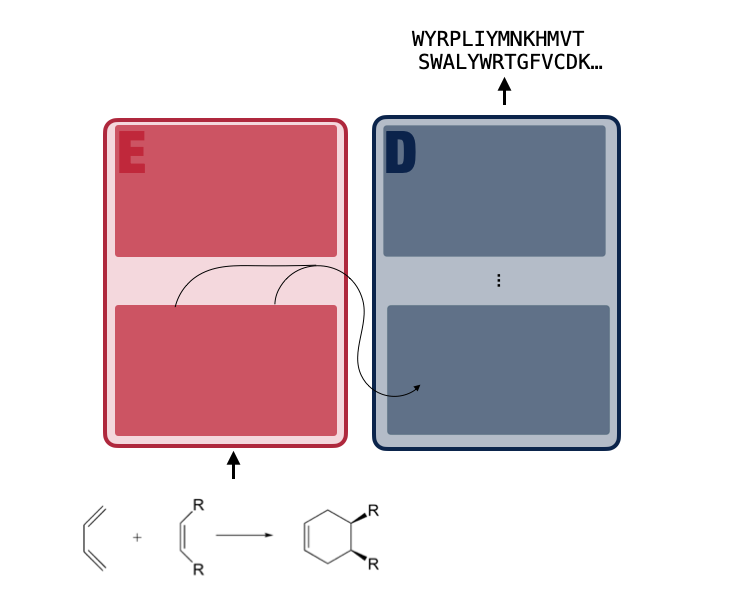
REXzyme (Reaction to Enzyme) (manuscript in preparation) is a translation machine -similar to Google Translator- for the generation of enzymes that catalize user-defined reactions.
It is possible to provide fine-grained input at the substrate level. Akin to how translation machines have learned to translate between complex language pairs with great success, often diverging in their representation at the character level (Japanese - English), we posit that an advanced architecture will be able to translate between the chemical and sequence spaces. REXzyme was trained on a set of 16,011 unique reactions and 20,911,485 enzyme pairs and it produces sequences that are predicted to perform their intended reactions.
you will need to provide a reaction in the SMILES format (Simplified molecular-input line-entry system). A useful online server to convert from molecules to SMILES can be found here: https://cactus.nci.nih.gov/chemical/structure.
After converting each of the reaction components you should convert them to canonical SMILEs using RDKit (https://www.rdkit.org/docs/GettingStartedInPython.html)
Finally, you should combine them in the following scheme: ReactantA.ReactantB>AgentA>ProductA.ProductB
e.g. for the carbonic anhydrase reaction: O=C([O-])O.[H+]>>O.O=C=O
We are still working in the analysis of the model for different tasks, including experimental testing. See below in this documentation information about the models' performance in different in-silico tasks and how to generate your own enzymes.
Model description
REXzyme is based on the Efficient T5 Large Transformer architecture (which in turn is very similar to the current version of Google Translator) and contains 48 (24 encoder/ 24 decoder) layers with a model dimensionality of 1024, totaling 770 million parameters.
REXzyme is a translation machine trained on portion the RHEA database containing 20,911,485 reaction-enzyme pairs. The pre-training was done on pairs of SMILES and amino acid sequences. Note that two seperate tokenizers were used for input (./tokenizer_aa-ABPE_SMILES/tokenizer_ABPE_rexzyme_offset) and labels (./tokenizer_aa).
REXzyme was pre-trained with a supervised translation objective i.e., the model learned to process the continous representation of the reaction from the encoder to autoregressively (causual language modeling) produce the output. The output tokens (amino acids) are generated one at a time, from left to right, and the model learns to match the original enzyme sequence. Hence, the model learns the dependencies among protein sequence features that enable a specific enzymatic reaction.
There are stark differences in the number of members among reaction classes. However, since we are tokenizing the reaction SMILES on a character level, the model has learnt dependencies among molecules and enzyme sequence features, and it can transfer learning from more to less populated reaction classes.
How to generate from REXzyme
REXzyme can be used with the HuggingFace transformer python package. Detailed installation instructions can be found here.
Since REXzyme has been trained on the objective of machine translation, users have to specify a chemical reaction, specified in the format of SMILES.
Disclaimer: Although the perplexity gets computed here it is not the best selection criteria. Usually the BLEU score is deployed for translation evaluation, but this score would enforce a high sequence similarity (thus not de novo design, which is what we tend to go for). We recommend generating many sequences and selecting them by plDDT, as well as other metrics.
Before running the inference script, one should create a text file containing the desired input SMILE. Note that if there are multiple reactions SMILE in the same file but in separate lines, the model will generate sequences for each reaction independently, creating different a different output file for each of them.
"""Inference on a SMILES txt. Saved as fastas
Previously called generate_comparison"""
if __name__ == '__main__':
from transformers import AutoTokenizer, AutoModelForSeq2SeqLM,AutoModelForCausalLM #T5ForConditionalGeneration
import argparse
import os
import torch
import json
parser = argparse.ArgumentParser(description='inference',
formatter_class=argparse.ArgumentDefaultsHelpFormatter)
parser.add_argument('--input_file', default='../inference/random_smiles2.txt', type=str,
help='File with the input molecule SMILES')
parser.add_argument('--model_path', default='./output03/checkpoint-60000', type=str, help='Path to model to load')
parser.add_argument('--tokenizer_aa',
default='/home/woody/b114cb/b114cb10/mol2pro/1.training-different-sizes/1.all-data-16M-tokenizernuria/tokenizer_aa', type=str,
help='Path to amino acid tokenizer')
parser.add_argument('--tokenizer_mol',
default='/home/woody/b114cb/b114cb10/mol2pro/1.training-different-sizes/1.all-data-16M-tokenizernuria/nuria_tokenizer_smiles', type=str,
help='Path to SMILES tokenizer')
parser.add_argument('--top_k',
default=15,type=int,
help='K for top-k sampling')
parser.add_argument('--output_folder', default='fastas', type=str, help='Folder for saving results')
args = parser.parse_args()
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
if 'gatgpt' in args.model_path.lower():
GNN = True
print('Graph data mode')
else:
GNN = False
print('SMILES/SELFIES data mode')
# Load protein tokenizer
if 'ape' in args.tokenizer_aa:
from ape_tokenizer import APETokenizer
tokenizer_aa = APETokenizer.from_pretrained(args.tokenizer_aa)
else:
tokenizer_aa = AutoTokenizer.from_pretrained(args.tokenizer_aa)
# Load molecule tokenizer
if GNN:
tokenizer_mol = None
else:
if 'ape' in args.tokenizer_mol:
from ape_tokenizer import APETokenizer
tokenizer_mol = APETokenizer.from_pretrained(args.tokenizer_mol)
else:
tokenizer_mol = AutoTokenizer.from_pretrained(args.tokenizer_mol)
# Load model
dec_only = False
if GNN:
from transformers import GPT2Config, Trainer
from models import GATGPT2Config, GATGPT2
from torch_geometric.data import Batch, Data
config = GATGPT2Config.from_pretrained(args.model_path)
# Load model weights
model = GATGPT2.from_pretrained(args.model_path, config=config)
model.eval()
model.to("cuda" if torch.cuda.is_available() else "cpu")
else:
try:
print('Attempt Seq2Seq model load... ')
model = AutoModelForSeq2SeqLM.from_pretrained(args.model_path).cuda()
except:
print('Attempt CausalLM model load... ')
model = AutoModelForCausalLM.from_pretrained(args.model_path).cuda()
model.config.eos_token_id = tokenizer_mol.eos_token_id
model.config.pad_token_id = tokenizer_mol.pad_token_id
print(
f"Set `eos_token_id` to {tokenizer_mol.eos_token_id} and `pad_token_id` to {tokenizer_mol.pad_token_id}.")
dec_only = True
print('Model Loaded')
smiles_list = []
with open(args.input_file, 'r') as input_file:
for line in input_file:
smiles_list.append(line.strip())
molecule_json = {}
for index,smiles in enumerate(smiles_list):
sequences=[]
if GNN:
from build_tokenized_dataset import convert_smiles_to_graph
node_feats, edge_index, edge_feats = convert_smiles_to_graph(smiles)
node_feats_tensor = torch.tensor(node_feats, dtype=torch.float, device=device)
edge_index_tensor = torch.tensor(edge_index, dtype=torch.long, device=device).T.contiguous()
edge_feats_tensor = torch.tensor(edge_feats, dtype=torch.float, device=device)
# Input to decoder is only bos
start_token = tokenizer_aa.bos_token_id or tokenizer_aa.convert_tokens_to_ids("▁") # fallback to the space which is always appended by our tokenizer
text_input_ids = torch.tensor([[start_token]], dtype=torch.long, device=device)
input_ids = {
"graph_node_feats": node_feats_tensor, # shape (N, 3)
"graph_edge_index": edge_index_tensor, # shape (2, E)
"graph_edge_feats": edge_feats_tensor, # shape (E, 2)
"batch": torch.full((len(node_feats),), 0, dtype=torch.long, device=device), # shape (N,)
"input_ids": text_input_ids
}
elif 'ape' in args.tokenizer_mol:
input_ids = tokenizer_mol(smiles, return_tensors="pt")["input_ids"].to(device='cuda')
else:
input_ids = tokenizer_mol(smiles, return_tensors="pt").input_ids.to(device='cuda')
if not GNN:
print(f'Generating for {smiles} (input ids: {input_ids})')
else:
print(f'Generating for {smiles}')
# top_k = Choose at random from the first K tokens (weigthed by softmax score)
# num_return_sequences = The number of independently computed returned sequences for each element in the batch.
if dec_only:
attention_mask = torch.ones_like(input_ids).cuda()
outputs = model.generate(input_ids, top_k=args.top_k, attention_mask = attention_mask, repetition_penalty=1.2, max_length=1024, do_sample=True, num_return_sequences=25)
else:
outputs = model.generate(input_ids, top_k=args.top_k, repetition_penalty=1.2, max_length=1024, do_sample=True, num_return_sequences=25)
sequences = [tokenizer_aa.decode(output, skip_special_tokens=True) for output in outputs]
if not os.path.exists(args.output_folder):
os.makedirs(args.output_folder)
filename = f'{args.output_folder}/output_topk{args.top_k}_file-{index}.fasta'
with open(filename, 'w') as fn:
for idx, seq in enumerate(sequences):
fn.write(f">{idx}\n{seq}\n")
# Store molecule name
molecule_json[filename] = smiles
# Save metadata
metadata_path = os.path.join(args.output_folder, 'molecule_input_metadata.json')
try:
with open(metadata_path, 'w') as json_file:
json.dump(molecule_json, json_file, indent=4)
print(f"Metadata successfully written to {metadata_path}")
except Exception as e:
print(f"An error occurred while writing to JSON: {e}")
- As a reference, the bash script should look something like this:
INFERENCE_FOLDER=output_folder # change to the name of the output folder you want
MODEL=checkpoint-90000 # path to the model
INFERENCE_TXT=reaction.txt # text file containing the reactions (in SMILE format) wanting to generate for.
source .environment/bin/activate # load an environment containing the required dependencies (transformers, torch, datasets)
python inference.py --input_file "$INFERENCE_TXT" --model_path "$MODEL" --output_folder "$INFERENCE_FOLDER" --tokenizer_mol tokenizer_ABPE_rexzyme_offset --tokenizer_aa tokenizer_aa
A word of caution
We have not yet fully tested the ability of the model for the generation of new-to-nature enzymes, i.e., with chemical reactions that do not appear in Nature (and hence neither in the training set). While this is the intended objective of our work, it is very much work in progress. We'll uptadate the model and documentation shortly.
Latest checkpoint of the model
Please use checkpoint-90000 for the latest parameters
- Downloads last month
- 3